pipelines-docs
Long-Read Oxford Nanopore Technologies (ONT)
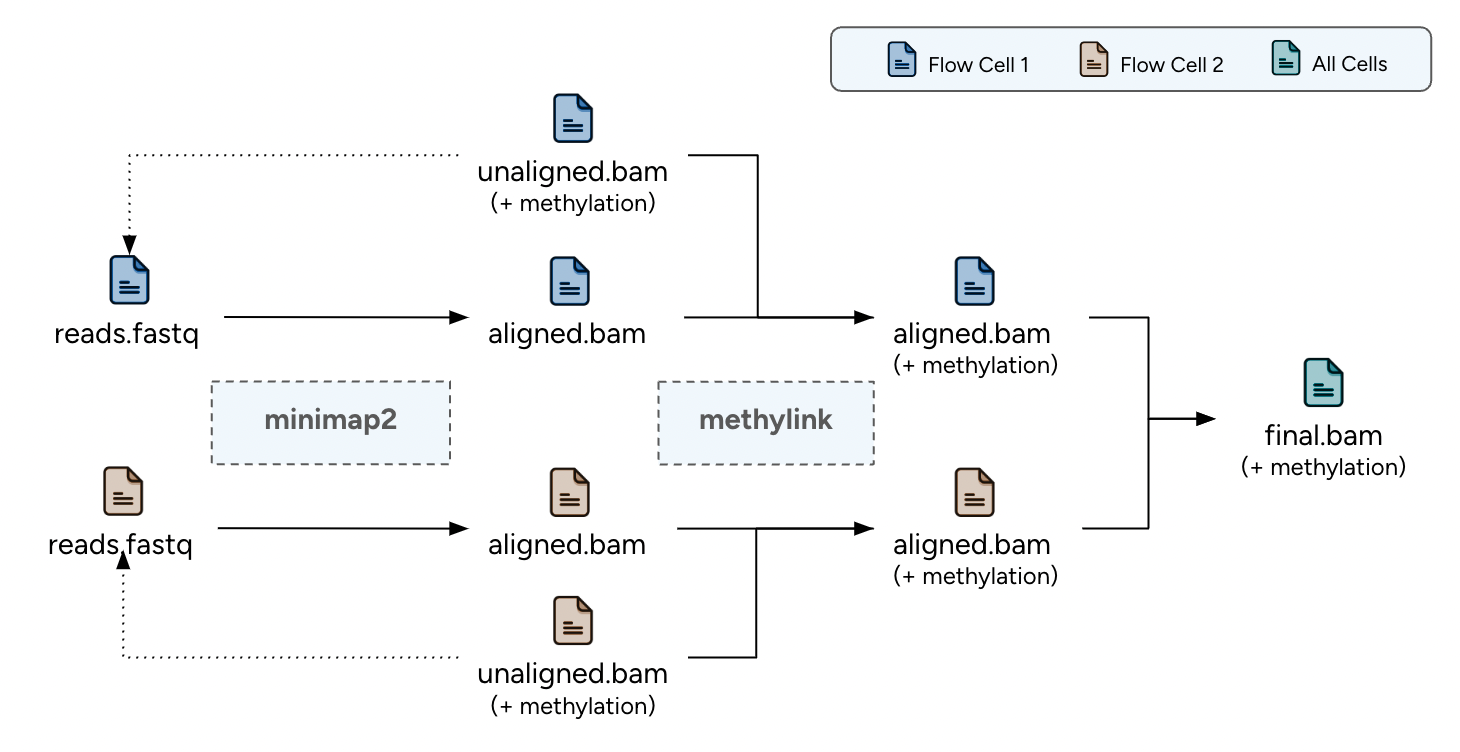
The long-read alignment pipeline for ONT data is designed for per-sample and per-library execution, handling one or multiple FASTQ files and the corresponding unaligned BAM files. The pipeline is optimized for distributed processing, requiring each FASTQ file to correspond to a single flow cell.
Key Pipeline Steps
- Alignment with minimap2: Initial alignment of the raw reads to the reference genome using minimap2.
- Read Groups Assignment: Assignment of reads to specific groups.
- Methylation and Tags Linking: Linking the methylation status information and other specific tags from the unaligned to the alignment BAM file.
Pipeline Chart