pipelines-docs
Long-Read RNA-seq, PacBio Kinnex
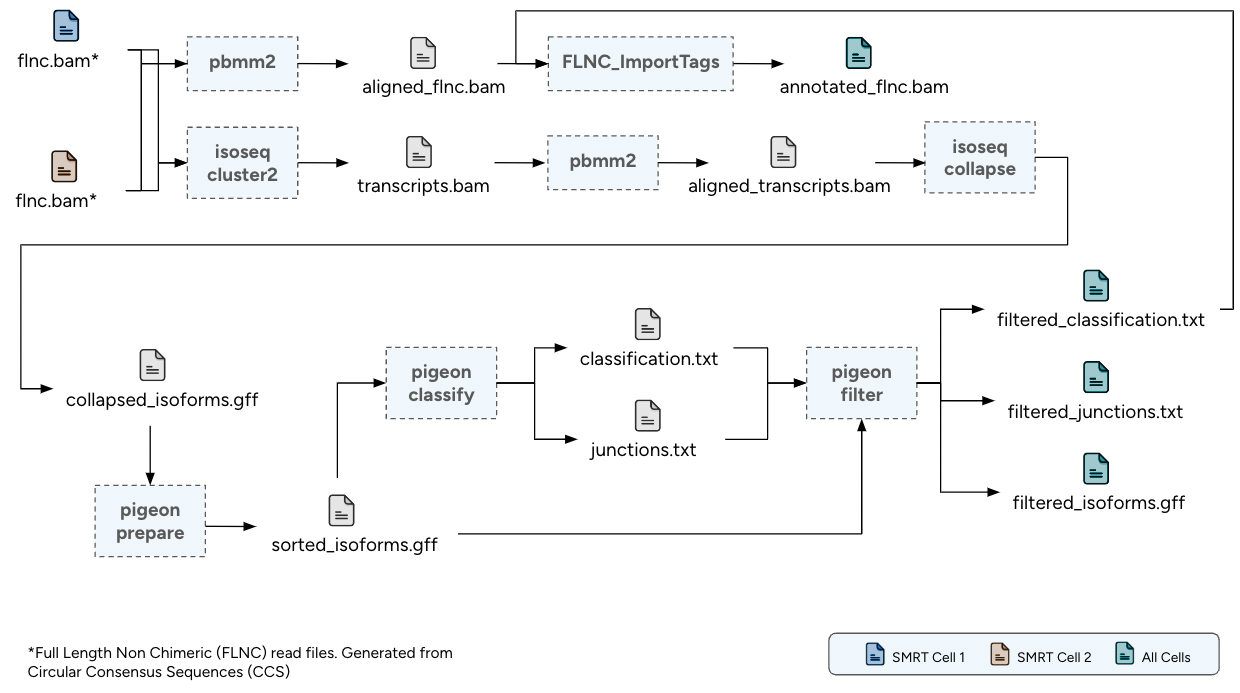
The long-read analysis pipeline for PacBio Kinnex RNA-seq data follows Iso-Seq processing guidelines1. It is designed for per-sample execution, handling one or more Full Length Non Chimeric (FLNC) BAM files. The pipeline has been extended to align and annotate FLNC reads with isoform-level information directly within the BAM file.
Key Pipeline Steps
- Read Clustering: Generation of high-quality consensus transcript sequences through clustering of FLNC reads.
- Alignment with pbmm2: Alignment of the consensus transcripts and FLNC reads to the reference genome using pbmm2.
- Transcript Collapsing: Collapsing of redundant transcripts based on exon-intron structure to define a unique set of isoforms.
- Isoform Classification and Filtering: Classification and filtering of isoforms to remove potential artifacts and retain high-confidence transcripts.
- Read Annotation: Annotation of aligned FLNC reads with isoform-level information embedded in the BAM file.
Pipeline Chart
1: PacBio Iso-Seq Analysis Guidelines. Available at: https://isoseq.how